
Affinity chromatography offers high selectivity, resolution, and capacity in most protein purification schemes. It has the advantage of utilizing a protein's biological structure or function for purification. As a result, purifications that would otherwise be time consuming and complicated, can often be easily achieved with affinity chromatography.

A commonly used metaphor to illustrate affinity binding is the lock and key analogy. A unique structure present on the surface of a protein is the key that will only bind to the corresponding lock, a specific ligand on a chromatographic support.
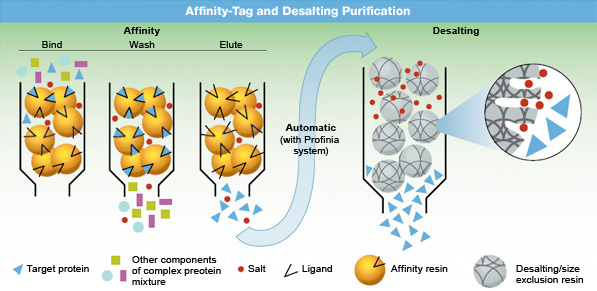
Affinity-tagged purification.
In two-step affinity-tagged protein purification, a protein is first purified by affinity chromatography, then desalted. In some medium pressure chromatography systems, such as the NGC medium pressure chromatography systems, these two steps can be automated. In the first step, a recombinant protein mixture is passed over a chromatography support containing a ligand that selectively binds proteins that contain an affinity-tag sequence (typically His or GST). Contaminants are washed away, and the bound protein is then eluted in pure form.
Affinity tags have different advantages. In immobilized metal affinity chromatography (IMAC), His binds with good selectivity to Ni2+ or other transition metals immobilized to the ligand; the tagged protein can be selectively eluted with imidazole. proteins tagged with GST bind to glutathione as the ligand, and are eluted with solutions of glutathione. Proteins with an enzymatically active GST fusion tag can only be purified under native conditions. In contrast, polyhistidine-tagged proteins may be purified under native or denaturing conditions.
During the second step of desalting, affinity-purified samples can simultaneously undergo buffer exchange to remove salts in preparation for downstream applications.
A number of desalting techniques, including size exclusion chromatography, dialysis, and ultrafiltration, also allow buffer exchange. Desalting often includes the removal not only of salt, but also of other foreign substances, such as detergents, nucleotides, and lipids.
Affinity chromatography can be broadly divided into two method types:
- The first method uses a naturally occurring structure or sequence of amino acids on the protein as the binding site. Examples include the affinity of Affi-Gel Blue support binding for albumin’s bilirubin-binding site and the binding of protein A in the Affi-Gel and Affi-Prep protein A supports to the Fc region of IgG. An important consideration for antibody purification is to determine the affinity of your target antibody for protein A/G chromatography media, which varies widely.
- The second method involves binding to a special amino acid sequence engineered into the protein of interest, commonly referred to as a "tag". A number of different tags are available. Two of the most commonly used protein tags are the polyhistidine tag, which binds to certain metal-containing complexes such as those in Profinity™ IMAC resins, and the glutathione s-transferase (GST) sequence, which binds to glutathione, found in Bio-Scale™ Mini Profinity™ GST media. Theoretically, any protein can be purified using the tagging method; however, many factors must be considered to design a process to purify tagged recombinant proteins.
Bio-Rad offers ready to use affinity media and customizable or activated media.
Ready to Use Affinity Media Selection Guide
| Ready-to-Use Affinity Media |
| | Matrix | Functional Group | Specificity | Capacity | Working pH | Pressure Limit | Applications |
| Nuvia™ IMAC | High capacity, pressure-stable polymer based on UNOsphere™ beads | NTA charged with Ni2+ | Histidine | ≥40 mg/ml* | 2–14 | 45 psi
(3.1 bar) | Purification of recombinant histidine-tagged proteins; can be charged with other transition metals |
| Profinity™ IMAC | Pressure-stable polymer based on UNOsphere beads | IDA, provided charged with Ni2+ and uncharged | Histidine | ≥15 mg/ml* | 1–14 | 100 psi
(6.8 bar) | Purification of recombinant proteins tagged with histidine; can be charged with other transition metals |
| Profinity™ GST** | Pressure-stable polymer based on UNOsphere beads | Immobilized glutathione | proteins tagged with GST | ≥10 mg/ml | 1–14 | 45 psi
(3.1 bar) | Purification of recombinant proteins tagged with GST |
| Profinity eXact™*** | Crosslinked 6% agarose | Subtilisin protease | Subtilisin prodomain | ≥3 mg/ml tag-free protein | 2–10 | 10 psi
(minus system pressure) | Generation of native, tag-free protein by on-column purification and cleavage |
| Affi-Gel®Protein A | Crosslinked agarose | Protein A
2 mg/ml | IgG | | 2–10 | 15 psi
(1 bar) | Purification of IgG from ascites, serum, and culture fluid; with MAPS buffer system, purification of 10 mg mouse IgG per ml of gel is possible |
| Affi-Prep®Protein A | Pressure-stable polymer | Protein A
2 mg/ml | IgG | | 2–10 | 1,000 psi
(70 bar) | Purified IgG from ascites, serum, and culture fluid; pressure-stable support for process-scale applications |
| Affi-Gel®Blue | Crosslinked agarose | Cibacron Blue F3GA 1.9 mg/ml | Albumin; general | ≥11 mg/ml | 2–10 | 15 psi
(1 bar) | Binds many nucleotide-requiring enzymes, albumin, and other proteins |
| DEAE Affi-Gel® Blue | Crosslinked agarose | Cibacron Blue F3GA and DEAE | Albumin and serum protiens | 0.14 ml serum/ml gel | 2–10 | 15 psi
(1 bar) | Purifies protease-free IgG from ascites, serum, and culture fluid with minimal sample preparation |
| CM Affi-Gel® Blue | Crosslinked agarose | Cibacron Blue F3GA and CM | Albumin and serum protiens | 0.17–0.5 ml serum/ml gel | 2–11 | 15 psi
(1 bar) | Produces albumin- and protease-free antibody preparation from serum without prior dialysis |
| Affi-Prep®Polymyxin | Pressure-stable polymer | Polymyxin
2–4 mg/ml | Endotoxins | >5 mg/ml | 2–10 | 1,000 psi
(70 bar) | Endotoxin removal |
| Affi-Gel®Boronate | Polyacrylamide gel | Boronate 1.05 ± 0.15 | cis-diols
meq/g | 130 µmol sorbitol/ml | 2–10 | 15 psi
(1 bar) | Adsorption of cis-hydroxyl–containing molecules, including sugars, nucleotides, and glycopeptides |
Activated media: Affinity chromatography can also include customized media. For instance, a specific ligand can be attached to an activated or coupled resin. Samples containing proteins that will bind to this ligand will be retained. One example is the attachment of DNA to beads via a coupling linkage. DNA-binding proteins (such as polymerases) will be retained on the media.
| Activated Media for Spontaneous Ligand Immobilization |
| | Matrix | Functional Group | Specificity | Capacity | Working pH | Pressure Limit | Applications |
| Profinity™ Epoxide | Pressure-stable polymer based on UNOsphere beads | Epoxy group | Nucleophiles; amini, thiol, -COOH | 36–40 mg/ml lgG | 1–14 | Up to 80 psi
(5.5 bar) | Activated matrix for the immobilization of various ligands (for example, protein A, StrepTactin, and immunoglobulins) |
| Affi-Gel®10 | Crosslinked agarose | N-hydroxy-succinimide ≥10 µmol/ml | -NH2 | 35 mg/ml | 3–11 | 15 psi
(1 bar) | For coupling proteins with pI 6.5–11 |
| Affi-Gel®15 | Crosslinked agarose | N-hydroxy-succinimide ≥9 µmol/ml | -NH2 | 35 mg/ml | 3–11 | 15 psi
(1 bar) | For coupling proteins with pI <6.5 |
| Affi-Gel®Hz | Crosslinked agarose | Hydrazide | Oxidized carbohydrates | 1–5 mg/ml | 2–10 | 15 psi
(1 bar) | Immobilization of immunoglobulins and other glycoproteins via carbohydrate molecules |
| Affinity Media Using Carbodiimide Activation |
| Affi-Gel®102 | Crosslinked agarose | -NH2 16 ± 4 meq/ml | -COOH | 40 mg | 2–11 | 15 psi
(1 bar) | Carbodiimide coupling of carboxyl-containing ligand |
* Refer to bulletin 3193 for purification conditions.
** For example, Profinity GST resin is only available in prepacked cartridges.
*** Profinity eXact Purification Resin.

The binding and elution conditions in affinity chromatography can vary greatly, however, there are some general guidelines. Because most binding interactions are based on those formed in nature, the conditions similar to those present in most cellular organisms are usually the best binding conditions. Therefore PBS (phosphate buffered saline) is often the buffer of choice. Conversely, conditions not normally found in vivo may alter the protein structure enough to cause the protein to dissociate from the ligand. In situations where there is no recommended elution condition, a low pH (>4) will often elute bound protein with an inherent risk of denaturation.